Page 243 - 2021_06-Haematologica-web
P. 243
Letters to the Editor
deletion. To this end, we reanalyzed the deleted sequences in the two cell lines. The original 3q21.3 frag- ment overlapping with LINC01565 relocated to the 3q26.2 site and deleted in the MOLM-1 clones displays high degree of conservation (Figure 3A, left panel), which might indicate functional importance of this sequence. ENCODE transcription factor (TF) ChIP data generated in K562 cells revealed a plethora of TF binding to this region (Online Supplementary Figure S2B). Some of these TF have predicted binding sites within the G2DHE sequence, including IKZF1, MAX, TAL1 and MAZ (Kiehlmeier et al., 2021, under review). Furthermore, EVI1 has been func-
tionally linked to some of these TF, including MTA1/2, HDAC1/2 and GATAD2B. All these proteins belong to the nucleosome remodeling and deacetylase (NuRD) complex,8 which was shown to specifically interact with the MDS-EVI1 (PRDM3) but not the EVI1 protein,9 while both EVI1 protein isoforms were shown to interact with HDAC1.10 Contrary to MOLM-1, no TF binding in the region deleted in the UCSD-AML clones was found (Online Supplementary Figure S2B).
Taken together, the observed reduction in EVI1 expres- sion upon breakpoint-RE deletion in MOLM-1 is more likely the consequence of TF binding loss within the
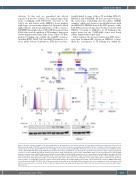
A
BC
D
Figure 2. Ectopic activation of EVI1 occurs via G2DHE, whereas breakpoint-associated retroelements do not display activating potential in the K562 cells. (A) Experimental strategy of CRISPR-Cas9-mediated genomic insertions. Donor templates containing either G2DHE (736 bp, red boxes) or 3q21.3 breakpoint-asso- ciated retroelements (breakpoint-RE) from selected 3q-AML cases (3071: 353 bp and MOLM-1: 280 bp, blue boxes) were inserted in the non-3q K562 reporter cell line (K562 eGFP-T2A-EVI1) using CRISPR-Cas9 downstream of EVI1 and within the last EVI1 intron. A genomic view shows the origin of 3q21.3 breakpoint sequences used in CRISPR experiments, dashed lines indicate 3q21.3 breakpoints in 3071 (left) and MOLM-1 (right). 3,071 homology-directed repair (HDR) template contains a part of MLT1J LTR element. G2DHE (red) and 3q21.3 breakpoint-RE samples (blue) are consistently colored throughout the figure. (B) Flow cytometry analysis on the K562 eGFP-T2A-EVI1 single clones bearing the desired G2DHE or 3q21.3 breakpoint insertions. Peaks corresponding to the green fluorescent protein (GFP) signal from single clones targeted at the same region are presented together on one graph, with peaks for untreated cells shown as a black outline. (C) Quantitative polymerase chain reaction (qPCR) analysis of the EVI1 mRNA levels in single clones shown in (B), relative to HMBS and normal- ized to the untreated eGFP-T2A-EVI1 cells. (D) Representative western blot of the full-length EVI1 isoform from the clones and untreated cells (ctr) shown in (B) and (C). Data shown in (C) are means of three technical replicates from one independent experiment. SINE: short interspersed nuclear elements; LTR: long ter- minal repeats.
haematologica | 2021; 106(8)
2271