Page 127 - 2021_06-Haematologica-web
P. 127
Regulation of F8 promoter activity
eliminated. Further shortening of the promoter (pF8-342) caused a minimal reduction of its activity with respect to pF8-1175, while a significant reduction (<20-40% pF8- 1,175) was observed with the pF8-246 (Figure 3C). Analysis of the shortened pF8 promoters in response to Ets overexpression, confirmed that pF8-600 and pF8-446 maintain the same response as pF8-1175, demonstrating a 3- and 5-fold upregulation with Ets-1 and Ets-1/Ets-2 com- bined (Figure 3D). Despite a clear reduction in its promot- er activity, pF8-342 interestingly maintained a ~3- to 5- fold activation with Ets-1 or Ets-1/Ets-2 combined, indi- cating that a specific response is maintained despite the elimination of one Ets-1 and one Ets-2 BS. When the pF8 was shortened further to 246 bp, a dramatic reduction in promoter activity was observed and was sustained despite Ets overexpression (Figure 3D). This was also validated in HEK293T cells (Online Supplementary Figure S1A and B). Overall, these results suggest that pF8 regulation may depend on the number of available Ets BS.
The second mutagenesis approach focused on the importance of each Ets site identified using a dissimilarity score <3. This approach selectively deleted the GGAA core from E1, E2, E3, E4, E5 Ets-1 and Ets-2 BSs (Figure 3B). As the Ets BSs are in the first 600 bp of pF8, we used pF8-600 to normalize the results as it preserves all the identified endothelial TFBS. Using this approach, we iden-
tified two important BS: -189 to -198 (E1) and -217 to -223 (E2). The disruption of E1 drastically reduced pF8 basal activity by approximately 60%, while the others failed to alter its activation (Figure 3E; Online Supplementary Figures S1C and S2A). Of note, the disruption of E1 site even pre- serving a response to Ets-1 overexpression abrogated the co-operative up-regulation mediated by Ets-1/2, thus agreeing with the elimination of the only Ets-2 site over- lapping that of Ets-1. On the other hand, disruption of the E2 site decreased exclusively the pF8 up-regulation in response to Ets overexpression (Figure 3F; Online Supplementary Figures S1D and S2B). Overall, the E1 and E2 sites appear to be central to pF8 regulation with E2 main- taining the basal pF8 activity in the absence of an Ets- induced upregulation.
Enhancement of F8 promoter activity by delivery of CRISPR/VPR activation system to E2 and E3 E26 transformation-specific binding sites
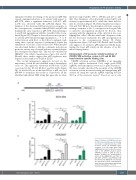
CRISPR activation system (CRISPRa) is an emerging tool that exploits deactivated Cas9, single guide RNA (sgRNA) and transcription activators for gene activation.34 We have recently described the potential of the CRISPR activation system in transactivating and upregulating pF8 activity by using two specific sgRNA targeting the first 300 bp of the promoter region.27 Based on our in silico
A
B
C
Figure 4. Delivery of CRISPRa to E2 and E3 E26 transformation-specific binding sites induces a significant increase in F8 promoter activity. (A) Schematic repre- sentation of the two single guide RNA (sgRNA) guides used to drive the recruit- ment of the CRISPR activation system (CRISPRa) system to pF8. (B and C) Graphs showing the transactivation effects of the sgRNA F8.1 and F8.2 on pF8-1175, pF8-600 and pF8-342 in (B) ECV-304 and (C) HEK293T cells. Results are expressed as mean ± standard devi- ation from two independent experiments performed in triplicate. *P<0.05
haematologica | 2021; 106(6)
1629