Page 245 - 2021_04-Haematologica-web
P. 245
Letters to the Editor
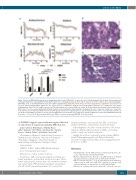
Figure 3. Effect of NFE2-K368X expression on megakaryopoiesis in vivo. FVB/N-45.1 acceptor mice were lethally irradiated (2x5 Gy given 4 hours apart) and transplanted with bone marrow (BM) from FVB/N-45.2 donor mice, lentivirally transduced either with an empty vector (n=5), or with a vector expressing wildtype (WT) NFE2 (n=6), or the NFE2-K368X mutant (n=6) (Online Supplementary Figure S4). Twelve weeks after the transplantation, engraftment exceeded 90% in all cases (Online Supplementary Figure S5). The expression level of NFE2 was doubled in transplanted BM compared to the endogenous level (Online Supplementary Figure S6). (A) Complete blood count. Mean±standard error of mean (SEM) are shown. (B) Representative Hematoxylin & Eosin (H&E) stained BM section of a mouse expressing the NFE2-K368X mutation, demonstrating the high variability in megakaryocyte size. *Large size; †middle size; ‡small size. Scale bar=50 mm. (C) H&E stained BM sections morphologically evaluated for megakaryopoiesis and enumerated for five mice of each genotype. Results rep- resent the mean+SEM per five HPF per mouse (400x magnification). Grouped data were analyzed for statistical significance by two-way ANOVA with Bonferroni post tests. ***P<0.001. (D) Representative BM section (H&E) of a mouse expressing NFE2-WT. Scale bar=50 mm.
or SUMO2/3 suggests a previously unrecognized function of sumoylation in negatively regulating NFE2 function.
Lukas Clemens Böckelmann,1 Titiksha Basu,1 Albert Gründer,1 Wei Wang,1 Jan Breucker,2 Sandra Kaiser,1 Andrea Pichler2 and Heike Luise Pahl1
1Department of Medicine I, Medical Center, University of Freiburg, Faculty of Medicine, University of Freiburg and 2Department of Epigenetics, Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany
Correspondence:
HEIKE L. PAHL - heike.pahl@uniklinik-freiburg.de doi:10.3324/haematol.2020.246587
Disclosures: no conflicts of interest to disclose.
Contributions: LCB conceived and designed experiments, per- formed experiments, analyzed data and wrote the paper; TB per- formed sequencing and analyzed data; AG performed experiments and analyzed data; WW performed experiments and analyzed data; JB and SK performed experiments; AP conceived and
designed experiments, and analyzed data; HLP conceived and designed experiments, analyzed data, and wrote the paper.
Acknowledgments: the authors sincerely thank Prof. Dr. Torsten Haferlach, Munich Leukemia Laboratory (MLL), for providing patients' samples and molecular diagnostics.
Funding: this work was supported by grants from the Mildred- Scheel-Doktorandenprogramm of the Deutsche Krebshilfe (70110768 to LCB) and by the Deutsche Forschungsgemeinschaft (Pa 611/5-3 to HLP).
References
1. McMullin MF, Cario H. LNK mutations and myeloproliferative dis- orders. Am J Hematol. 2016;91(2):248-251.
2. Rumi E, Harutyunyan AS, Pietra D, et al. LNK mutations in familial myeloproliferative neoplasms. Blood. 2016;128(1):144-145.
3. Oh ST, Simonds EF, Jones C, et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116(6):988-992.
4. Wiestner A, Schlemper RJ, van der Maas APC, Skoda RC. An acti-
haematologica | 2021; 106(4)
1161
A
B
C
D