Page 243 - 2021_04-Haematologica-web
P. 243
Letters to the Editor
SUMO2/3.14 PIAS1 is also involved in the negative regu- lation of the JAK/STAT signaling pathway,12 which is aberrantly upregulated in MPN patients and constitutes a central molecular hallmark of MPN pathogenesis.
Negative regulation of transcription factors by sumoy- lation has previously been described and can occur through several distinct mechanisms.15 For example, sumoylation can compete with acetylation or phosphory- lation of nearby residues. While the acetylated or phos- phorylated transcription factor is active, the sumoylated form is not. NFE2 activity is regulated by phosphoryla- tion, but the precise molecular mechanisms have not been defined.9 We, therefore, propose that sumoylation
may negatively regulate NFE2 by interfering with other post-translational modifications, such as acetylation, methylation or phosphorylation, required for full activity.
Alternatively, rather than controlling the placement of other post-transcriptional modifications, NFE2 sumoyla- tion may regulate nuclear localization. PIAS1 and ZNF451-N sumoylate the tumor suppressor PML, the main component of PML nuclear bodies (PML-NBs).12 PML-NB control gene expression by sequestering tran- scriptions factors in a SUMO-dependent manner.15 WT-NFE2 and PML co-localize in the nucleus of K562 cells, while sumoylation-deficient NFE2 did not.10 Sumoylated WT-NFE2 might, therefore, become
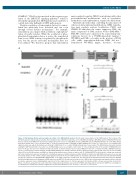
Figure 1. DNA binding affinity and transcriptional activity of the NFE2-K368X mutant. (A) Schematic representation of the NFE2 protein. The location of the K368X mutation is indicated by an asterisk, the sumoylation site at K368 is marked by a circle. (B) EMSA of wild-type (WT) NFE2 and the NFE2-K368X mutant. Nuclear extracts from HEK293T cells transduced with expression vectors encoding MafG as well as NFE2-WT (lane 3), NFE2-K368X (lane 8), or the NFE2-262aa truncation mutant (lane 7) were incubated with a 32P-labeled oligonucleotide containing an NFE2 binding site. In lane 4, a 100x excess of a nonradioactive oligonucleotide was added. Alternatively, an antibody to NFE2 (lane 5) or a control NF-κB antibody (lane 6) was added. The NFE2-262aa truncation mutant, lacking the bZIP domain with consecutive loss of DNA binding, serves as a negative control (lane 7).6 The arrowhead points to the position of NFE2/DNA com- plexes on the gel, the open circle indicates nonspecific bands. (C) Dual luciferase reporter assay. HEK293T cells were co-transfected with a pRBGP2-luciferase reporter construct that contains tandemly arranged NFE2 binding sites driving a minimal chicken β-globin promoter together with expression vectors for NFE2, either WT or the K368X mutant, and MafG as indicated. Experiments were carried out with a ratio of 1:8 MafG:NFE2. Firefly luciferase activity was measured 24 h after transfection and was normalized to constitutively expressed renilla luciferase activity. Activity for transfection with MafG alone was set as one and fold activity relative to this control is depicted. Bar graphs represent the mean + SEM. (D) Rescue of β-globin expression. CB3 cells were infected with lentiviral (pLeGO-iG) constructs encoding NFE2-WT, NFE2-K368X, or an empty control virus as indicated. 72 h after infection, RNA was harvested and assayed for β-globin and β2-microglobulin housekeeping gene mRNA expression by qRT-PCR. Results represent the mean + SEM of three independent experiments and are reported as relative expression levels setting β-globin expression for the empty virus as 1. Protein expression in transduced CB3 cells was assessed by western blot. Whole cell extracts, prepared from each of the three independent experiments, were probed for NFE2 and stripped blots reprobed for GAPDH as a loading con- trol. All data were analyzed for statistical significance by two-tailed Student’s t-test. *P<0.05; **P<0.01; ****P<0.0001.
haematologica | 2021; 106(4)
1159
A
B
C
D