Page 161 - 2020_11-Haematologica-web
P. 161
Letters to the Editor
AB
CD
E
F
F
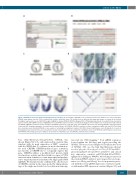
Figure 3. NPAS4L is involved in chicken hemangioblast specification. (A) Screenshot of NPAS4L locus in chicken promoterome database (see text for web link). NPAS4L transcription start site (TSS) is indicated by red label and black arrow. TSS activity levels at different developmental stages are shown at the bottom. The highest expression is seen at HH7. (B) Design of sgRNA for chicken NPAS4L CRISPRa. Mapped TSS is TCAGCAGG (underlined). Preceding 500 bp of promoter region is shown, with sgRNA sequences highlighted in red and PAM sequences in blue. (C) Schematic diagram of how embryos are electroporated and cultured. (D) NPAS4L CRISPRa constructs activate endogenous NPAS4L expression ectopically (oval). (Top) NPAS4L expression only; (bottom) NPAS4L expression togeth- er with anti-GFP staining marking the electroporated territories (brown). (E) NPAS4L CRISPRa constructs activate endogenous LMO2 (left) and SCL/TAL1 (mid- dle) ectopically (oval). Zebrafish NPAS4L is also capable of activating hemangioblast markers (SCL/TAL1 shown in right panel, oval). (F) Hypothetic scenario of hemangioblast specification in the ancestral amniote and ancestral reptile. It is proposed that the zebrafish scenario (both NPAS4L and ETV2 genes are present, with NPAS4L functioning upstream of ETV2) is the default one. Mammals have lost NPAS4L and birds have lost ETV2.
base (http://fantom.gsc.riken.jp/zenbu/), NPAS4L was shown (Figure 3A) to be only expressed in a narrow time window with its peak expression at HH7, consistent with the WISH data. To evaluate its molecular function, we used CRISPRa (CRISPR-mediated gene activation; also known as CRISPR-on)23 to ectopically express this gene. CRISPRa utilizes a modified Cas9 protein (with dead nuclease activity and fused with ten copies of VP16 transactivation domain) to recruit transcriptional machin- ery to targeted promoters mediated by single guide RNA (sgRNA). We had previously confirmed the effectiveness of CRISPRa system in the avian model by taking advan- tage of the single-nucleotide level resolution in transcrip-
tion start site (TSS) mapping.22 Four sgRNA sequences located within the 500-base pair region preceding the NPAS4L TSS were selected (Figure 3) (for interactive view of NPAS4L TSS, use the link http://fantom.gsc.riken.jp/ zenbu/gLyphs/#config=b1zZI1gUFZ6mHX6- 4Gvxr;loc=galGal5::chr3:4300587..4304021+) and cloned into expression construct pAC154-dual-dCas9VP160- sgExpression (Addgene #48240). Mesoderm precursors in the streak in HH2/3 embryos were targeted for electropo- ration (see Weng and Sheng19 for electroporation proto- col) with these four sgRNA expression constructs togeth- er with marker GFP expression construct (Figure 3C), and
haematologica | 2020; 105(11)
2649