Page 148 - Haematologica - Vol. 105 n. 6 - June 2020
P. 148
S. Ferrero et al.
Validation set
The Nordic Lymphoma Group MCL2 and MCL3, phase 2, prospective trials17 were used for independent validation of our findings. In particular, the raw sequencing data of the study by Eskelund et al. were reanalyzed according to our bioinformatics pipeline (detailed before), to get a uniform mutation calling.
Results
Patients characteristics
Out of the 300 patients enrolled in the FIL-MCL0208 clinical trial, 186 (62%) were provided with CD19+ sorted tumor cells from the BM and were evaluable for both mutations and copy number abnormalities. Moreover, four more patients were provided with the copy number
abnormalities data only. Baseline features of the cases included in the molecular study overlapped with those cases not included in the molecular analysis because of a lack of tumor material in the BM aspirates. As expected, tumor cells were obtained more frequently in cases with BM infiltration documented by morphological or flow- cytometry analysis (Table 1). Overall, this observation did not introduce a selection bias, since cases evaluable for genomic studies showed a similar outcome to cases not analyzed, both in terms of PFS and OS (Online Supplementary Figure S1).
Description of genomic alterations
At least one somatic non-synonymous mutation affect- ing genes of the target region was observed in 69.8% of patients (130 of 186) (Figure 1, Online Supplementary Figure
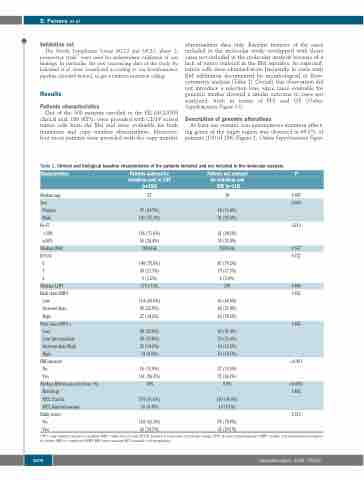
Table 1. Clinical and biological baseline characteristics of the patients included and not included in the molecular analysis.
Characteristics
Median age
Sex Female Male
Ki-67 <30% ≥30%
Median WBC ECOG
0 1 2
Median LDH Risk class MIPI
Low Intermediate High
Risk class MIPI-c
Low Low-Intermediate Intermediate/High High
BM invasion No
Yes
Median BM invasion by flow (%)
Histology
MCL Classic
MCL blastoid variant
Bulky mass No
Yes
Patients analysed for mutations and/or CNV (n=190)
57
47 (24.7%)
143 (75.3%)
126 (71.6%) 50 (28.4%) 74500/uL
144 (75.8%) 40 (21.1%) 6 (3.2%) 275.5 UI/L
114 (60.0%) 49 (25.8%) 27 (14.2%)
88 (50.0%) 49 (27.8%) 25 (14.2%) 14 (8.0%)
26 (13.9%) 161 (86.1%) 10%
174 (91.6%)
16 (8.4%) 124 (65.3%)
66 (34.7%)
Patients not analysed for mutations and
CNV (n=110)
58
18 (16.4%)
92 (83.6%)
61 (64.2%) 34 (35.8%) 75000/uL
87 (79.2%) 19 (17.3%) 4 (3.6%) 298
66 (60.0%) 24 (21.8%) 20 (18.2%)
45 (47.4%) 30 (31.6%) 10 (10.5%) 10 (10.5%)
37 (33.9%) 72 (66.1%) 0.8%
100 (90.9%)
10 (9.1%) 78 (70.9%)
32 (29.1%)
P
0.987
0.090
0.210
0.567 0.722
0.848 0.562
0.685
<0.001
<0.0001
0.842
0.315
CNV: copy number variation analysis;WBC: white blood cells; ECOG: Eastern Cooperative Oncology Group; LDH: lactate dehydrogenase; MIPI: mantle cell international prognos- tic index; MIPI-c: combined MIPI; BM: bone marrow. MCL: mantle cell lymphoma.
1606
haematologica | 2020; 105(6)