Page 127 - Haematologica - Vol. 105 n. 6 - June 2020
P. 127
PTCL classification using RT-MLPA assay
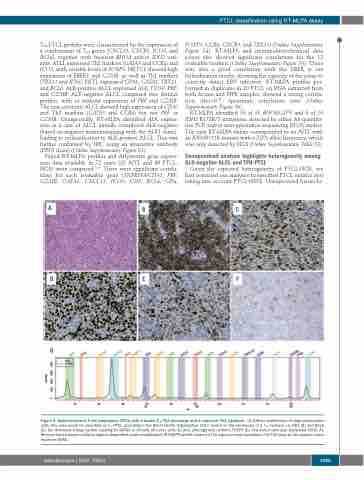
TFH-PTCL profiles were characterized by the expression of a combination of TFH genes (CXCL13, CXCR5, ICOS, and BCL6), together with frequent RHOA and/or IDH2 vari- ants. ATLL expressed Th2 markers (GATA3 and CCR4) and ICOS, with variable levels of FOXP3. NKTCL showed high expression of EBER1 and GZMB, as well as Th1 markers (TBX21 and IFNγ). HSTL expressed CD56, GATA3, TBX21, and BCL6. ALK-positive ALCL expressed ALK, CD30, PRF, and GZMB. ALK-negative ALCL comprised two distinct profiles, with or without expression of PRF and GZMB. The non-cytotoxic ALCL showed high expression of CD30 and Th2 markers (GATA3 and CCR4) but not PRF or GZMB. Unexpectedly, RT-MLPA identified ALK expres- sion in a case of ALCL initially considered ALK-negative (based on negative immunostaining with the ALK1 clone), leading to reclassification to ALK-positive ALCL. This was further confirmed by IHC using an alternative antibody (D5F3 clone) (Online Supplementary Figure S3).
Paired RT-MLPA profiles and Affymetrix gene expres- sion data available in 72 cases (23 AITL and 49 PTCL- NOS) were compared.18,19 There were significant correla- tions for each evaluable gene (TNFRSF8/CD30, PRF, GZMB, GATA3, CXCL13, ICOS, CD8, BCL6, CD4,
FOXP3, CCR4, CXCR5, and TBX21) (Online Supplementary Figure S4). RT-MLPA and immunohistochemical data scores also showed significant correlations for the 12 evaluable markers (Online Supplementary Figure S5). There was also a good correlation with the EBER in situ hybridization results, showing the capacity of the assay to correctly detect EBV infection. RT-MLPA profiles per- formed in duplicates in 20 PTCL on RNA extracted from both frozen and FFPE samples, showed a strong correla- tion (rho>0.7, Spearman correlation test) (Online Supplementary Figure S6).
RT-MLPA identified 33 of 33 RHOAG17V and 9 of 10 IDH2 R172K/T mutations, detected by either AS-quantita- tive PCR and/or next-generation sequencing (NGS) studies. The only RT-MLPA failure corresponded to an AITL with an IDH2R172K mutant with a 2.8% allele frequency, which was only detected by NGS (Online Supplementary Table S2).
Unsupervised analysis highlights heterogeneity among ALK-negative ALCL and TFH-PTCL
Given the expected heterogeneity of PTCL-NOS, we first restricted our analyses to specified PTCL entities (not taking into account PTCL-NOS). Unsupervised hierarchi-
A
BC
DEF
G
Figure 2. Nodal peripheral T-cell lymphomas (PTCL) with a double TFH/Th2 phenotype and a molecular Th2 signature. (A) Diffuse proliferation of large pleomorphic cells; this case would be classified as TFH PTCL according to the World Health Organization 2017, based on the expression of 2 TFH markers, i.e. PD1 (B) and BCL6 (C), but disclosed strong nuclear staining for GATA3 in virtually all tumor cells (D) and, although less uniform, FOXP3 (E). Few tumor cells also expressed CD30 (F). Reverse transcriptase-multiplex ligation-dependent probe amplification (RT-MLPA) profile showed a Th2 signature and classified in the Th2 class by the support vector machine (SVM).
haematologica | 2020; 105(6)
1585