Page 171 - Haematologica May 2020
P. 171
Ph– ALL cytogenetics and allogeneic HCT outcomes
t(4;11)(q21;q23), complex karyotype, t(8;14)(q24;q32), low hypodiploidy, or near triploidy, among others.1 However, only a subset of Ph– patients underwent allogeneic HCT in those trials. Thus, the applicability of existing cytogenetic risk classifications for allogeneic transplant recipients with ALL remains uncertain due to the relatively low frequency of specific Ph– cytogenetic abnormalities and the modest size of prior studies. In a single-center retrospective cohort study of 333 allograft recipients with ALL, cytogenetic risk did not predict survival after allogeneic HCT.2 Notably, in that study Ph+ patients accounted for the majority of patients in the poor-risk cytogenetic group, and the cyto- genetic risk scheme used was chosen arbitrarily. Another study on allogeneic HCT in Ph– ALL (n=373), conducted in Japan, found no difference in overall survival between patients with high-risk [t(4;11), t(8;14), low hypodiploidy, and complex karyotype] and standard-risk cytogenetics.3 A more recent analysis of Ph– B-ALL patients from GRAALL clinical trials identified t(4;11)/KMT2A-AFF1 and t(v;14q32)/IGH as markers of poor clinical outcome; how- ever, only a third of the trial patients underwent allogeneic HCT in first complete remission.4
In view of the conflicting prior data, we analyzed Center for International Blood and Marrow Transplant Research (CIBMTR) registry data to determine the prog- nostic impact of individual conventional (G-banding)
cytogenetic abnormalities and major previously estab- lished Ph– cytogenetic risk classifications (Table 1) on outcomes of allogeneic HCT. We also developed an allo-
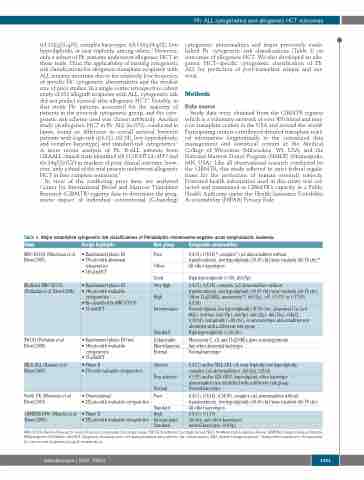
Table 1. Major established cytogenetic risk classifications of Philadelphia chromosome-negative acute lymphoblastic leukemia.
geneic HCT‐specific cytogenetic classification of Ph ALL for prediction of post-transplant relapse and sur- vival.
Methods
Data source
Study data were obtained from the CIBMTR registry which is a voluntary network of over 450 blood and mar- row transplant centers in the USA and around the world. Participating centers contributed detailed transplant-relat- ed information longitudinally to the centralized data management and statistical centers at the Medical College of Wisconsin (Milwaukee, WI, USA) and the National Marrow Donor Program (NMDP) (Minneapolis, MN, USA). Like all observational research conducted by the CIBMTR, this study adhered to strict federal regula- tions for the protection of human research subjects. Protected health information used in this study was col- lected and maintained in CIBMTR’s capacity as a Public Health Authority under the Health Insurance Portability Accountability (HIPAA) Privacy Rule.
–
Study
MRC-ECOG (Moorman et al. Blood 2007)
Modified MRC-ECOG (Pullarkat et al. Blood 2008)
SWOG (Pullarkat et al. Blood 2008)
NILG-ALL (Bassan et al. Blood 2009)
North UK (Moorman et al. Blood 2010)
GIMEMA 0496 (Mancini et al. Blood 2005)
Design highlights
• Randomized phase III • 796 pts with abnormal
cytogenetics • 310 alloHCT
• Randomized phase III • 140 pts with evaluable
cytogenetics
• Re-classified by MRC-ECOG • 19 alloHCT
• Randomized phase III trial • 140 pts with evaluable
cytogenetics • 19 alloHCT
• Phase II
• 276 with evaluable cytogenetics
• Observational
• 292 pts with evaluable cytogenetics
• Phase II
• 282 pts with evaluable cytogenetics
Risk group
Poor
Cytogenetic abnormalities
t(4;11), t(8;14)*, complex* (≥5 abnormalities without
translocations), low hypodiploidy (30-39 chr)/near triploidy (60-78 chr)* All other karyotypes
Monosomy 7, +8, and 11q23/MLL gene rearrangements Any other abnormal karyotype
Normal karyotype
Other
Good
Very high High Intermediate
Standard Unfavorable
Miscellaneous
Normal
Adverse
Non-adverse
Normal Poor
High hyperdiploidy (>50), del(9p)
t(4;11), t(8;14), complex (≥5 abnormalities without
translocations), low hypodiploidy (30-39 chr)/near triploidy (60-78 chr) Other 11q23/MLL, monosomy 7§, del(7p), +8§, t(1;19) or t(17;19), t(5;14)
Normal diploid, low hyperdiploidy (47-50 chr), abnormal 11q (not MLL), del(6q), del(17p), del(9p), del(12p), del(13q), t14q32,
t(10;14), tetraploidy (>80 chr), or any karyotype abnormalities not identified with a different risk group
High hyperdiploidy (>50 chr)
t(4;11) and/or MLL-AF4, +8, near triploidy, low hypodiploidy, complex (≥3 abnormalities), del(6q), t(8;14)
t(1;19) and/or E2A-PBX1, hyperdiploid, other karyotype abnormalities not identified with a different risk group Normal karyotype
Standard
High Intermediate Standard
t(4;11), t(8;14), t(14;18), complex (≥5 abnormalities without translocations), low hypodiploidy (30-39 chr)/near triploidy (60-78 chr) All other karyotypes
t(4;11), t(1;19)
del(6q) and other karyotypes
normal karyotype, del(9p)
MRC-ECOG: Medical Research Council-Eastern Cooperative Oncology Group; SWOG: Southwest Oncology Group; NILG: Northern Italy Leukemia Group; GIMEMA: Gruppo Italiano Malattie EMatologiche dell'Adulto; alloHCT: allogeneic hematopoietic cell transplantation; pts: patients; chr: chromosomes; MLL: mixed lineage leukemia *Independent predictors. §Unfavorable by Cancer and Leukemia Group B classification.
haematologica | 2020; 105(5)
1331