Page 110 - Haematologica May 2020
P. 110
M.N. Peiris et al.
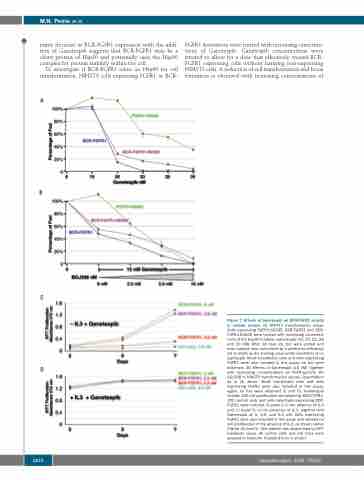
matic decrease in BCR-FGFR1 expression with the addi- tion of Ganetespib suggests that BCR-FGFR1 may be a client protein of Hsp90 and potentially uses the Hsp90 complex for protein stability within the cell.
To investigate if BCR-FGFR1 relies on Hsp90 for cell transformation, NIH3T3 cells expressing FGFR1 or BCR-
A
FGFR1 derivatives were treated with increasing concentra- tions of Ganetespib. Ganetespib concentrations were titrated to allow for a dose that effectively treated BCR- FGFR1 expressing cells without harming non-expressing NIH3T3 cells. A reduction of cell transformation and focus formation is observed with increasing concentrations of
B
C
D
Figure 7. Effects of Ganetespib on BCR-FGFR1 activity in cellular assays. (A) NIH3T3 transformation assay. Cells expressing FGFR1-K656E, BCR-FGFR1 and BCR- FGFR1-K656E were treated with increasing concentra- tions of the Hsp90 inhibitor, Ganetespib (10, 20, 23, 26 and 30 nM). After 14 days (d), foci were scored and each sample was normalized by transfection efficiency set to 100% as the starting value under conditions of no Gantespib. Mock transfected cells and cells expressing FGFR1 were also included in this assay; no foci were observed. (B) Effects of Ganetespib (15 nM) together with increasing concentrations of FGFR-specific TKI BGJ398 in NIH3T3 transformation assays. Quantitation as in (A) above. Mock transfected cells and cells expressing FGFR1 were also included in this assay; again, no foci were observed (C and D). Ganetespib inhibits 32D cell proliferation stimulated by BCR-FGFR1. 32D control cells and cells selectively expressing BCR- FGFR1 were cultured in panel C in the absence of IL-3 and, in panel D, in the presence of IL-3, together with Ganetespib at 0, 2.5, and 5.0 nM. Cells expressing FGFR1 were also included in this assay and showed no cell proliferation in the absence of IL-3, as shown earlier (Figure 3A and D). Cell viability was determined by MTT metabolic assay. All control cells and cell lines were assayed in triplicate. Standard error is shown.
1270
haematologica | 2020; 105(5)