Page 104 - Haematologica May 2020
P. 104
M.N. Peiris et al.
AB
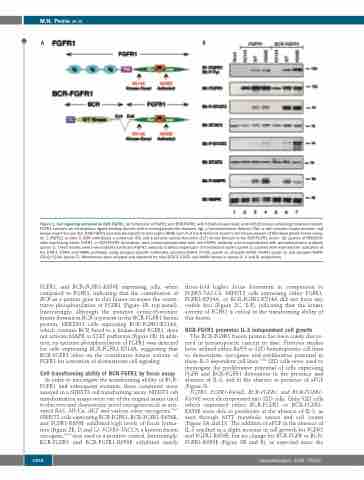
Figure 1. Cell signaling activated by BCR-FGFR1. (A) Schematic of FGFR1 and BCR-FGFR1 with K514A kinase dead, and K656E kinase activating mutations shown. FGFR1 contains an extracellular ligand binding domain with immunoglobulin-like domains (Ig), a transmembrane domain (TM), a split tyrosine kinase domain, and kinase insert domain (KI). BCR-FGFR1 contains breakpoint cluster region (BCR) exon 4 at the N-terminus fused to the kinase domain of fibroblast growth factor recep- tor 1 (FGFR1) at exon 9. BCR contributes a coiled-coil (CC) and a putative serine/threonine (S/T) kinase domain to the BCR-FGFR1 fusion. (B) Lysates of HEK293T cells expressing either FGFR1 or BCR-FGFR1 derivatives were immunoprecipitated with anti-FGFR1 antibody and immunobotted with phosphotyrosine antibody (panel 1). These lysates were immunoblotted with anti-FGFR1 antibody to detect expression of transfected clones (panel 2). Lysates were examined for activation of the STAT3, STAT5 and MAPK pathways using phospho-specific antibodies; phospho-STAT3 (Y705) (panel 3), phospho-STAT5 (Y694) (panel 5) and phospho-MAPK (T202/Y204) (panel 7). Membranes were stripped and reprobed for total STAT3, STAT5 and MAPK shown in panels 4, 6 and 8, respectively.
FGFR1, and BCR-FGFR1-K656E expressing cells, when compared to FGFR1, indicating that the contribution of BCR as a partner gene to this fusion increases the consti- tutive phosphorylation of FGFR1 (Figure 1B, top panel). Interestingly, although the putative serine/threonine kinase domain in BCR is present in the BCR-FGFR1 fusion protein, HEK293T cells expressing BCR-FGFR1-K514A, which contains BCR fused to a kinase-dead FGFR1, does not activate MAPK or STAT pathways (Figure 1B). In addi- tion, no tyrosine phosphorylation of FGFR1 was detected for cells expressing BCR-FGFR1-K514A, suggesting that BCR-FGFR1 relies on the constitutive kinase activity of FGFR1 for activation of downstream cell signaling.
Cell transforming ability of BCR-FGFR1 by focus assay
In order to investigate the transforming ability of BCR- FGFR1 and subsequent mutants, these constructs were assayed in a NIH3T3 cell transforming assay. NIH3T3 cell transformation assays were one of the original assays used to discover and characterize novel oncogenes such as acti- vated RAS, MUC4, AKT and various other oncogenes.16,17 NIH3T3 cells expressing BCR-FGFR1, BCR-FGFR1-K656E, and FGFR1-K656E exhibited high levels of focus forma- tion (Figure 2B, D and G). FGFR3-TACC3, a known fusion oncogene,15,16 was used as a positive control. Interestingly, BCR-FGFR1 and BCR-FGFR1-K656E exhibited nearly
three-fold higher focus formation in comparison to FGFR3-TACC3. NIH3T3 cells expressing either FGFR1, FGFR1-K514A, or BCR-FGFR1-K514A did not form any visible foci (Figure 2C, E-F), indicating that the kinase activity of FGFR1 is critical to the transforming ability of this fusion.
BCR-FGFR1 promotes IL-3 independent cell growth
The BCR-FGFR1 fusion protein has been solely discov- ered in hematopoietic cancers to date. Previous studies have utilized either Ba/F3 or 32D hematopoietic cell lines to demonstrate oncogenic and proliferative potential in these IL-3 dependent cell lines.16,18 32D cells were used to investigate the proliferative potential of cells expressing FGFR and BCR-FGFR1 derivatives in the presence and absence of IL-3, and in the absence or presence of aFGF (Figure 3).
FGFR1, FGFR1-K656E, BCR-FGFR1, and BCR-FGFR1- K656E were electroporated into 32D cells. Only 32D cells which expressed either BCR-FGFR1 or BCR-FGFR1- K656E were able to proliferate in the absence of IL-3, as seen through MTT metabolic assays and cell counts (Figure 3A and D). The addition of aFGF in the absence of IL-3 resulted in a slight increase in cell growth for FGFR1 and FGFR1-K656E, but no change for BCR-FGFR or BCR- FGFR1-K656E (Figure 3B and E), as expected since the
1264
haematologica | 2020; 105(5)