Page 70 - Haematologica April 2020
P. 70
P. Balogh et al.
transcriptional programs affected have been previously linked to RUNX3 function, including quiescence, DNA- damage responsiveness, and TGFβ signaling.12,20,26,36,37 Decreased RUNX3 in aged tissues has been previously reported but was analyzed in heterogeneous mixtures of mature cell types.15-18 Our results derive from purified, long-lived stem cells and reveal conservation between mice and humans. The repressive mechanism likely relates to the aging-associated epigenetic changes identi- fied. Beerman et al. have shown that murine HSC aging and proliferative stress reconfigure the DNA methylation landscape, with key erythroid and lymphoid regions tar- geted for hyper-methylation and repression.6 The increased Runx3 P2 promoter methylation we identified in aged murine HSC has also been found in aging of other tissues and cancers.16,27,38 The aging-associated decreases in H3K27ac that we identified at the human RUNX3 pro- moter and upstream super-enhancer may contribute to its repression in human HSC.
A feature of HSC aging conserved from mice to humans consists of myeloid skewing characterized by augmented marrow production of neutrophils and mono- cytes at the expense of erythroid and lymphoid cells.1-3 RUNX3 deficiency in aged mice likewise yields increased myelopoiesis; however, it presents as a myeloprolifera- tive disorder, which appears to be distinct from age-asso- ciated myeloid skewing due to lack of B-cell and T-cell perturbations, and a relatively mild erythroid deficit that may be secondary to leukocytosis.22 Despite these differ-
ences, our study indicates that loss of RUNX3 diminishes the expression of genes required for the erythroid pro- gram, and potentially de-represses myeloid genes. Epigenetic and transcriptomic analysis of aged HSC have also indicated that myeloid-skewing may manifest after similar changes.6,7 Other aging-like phenotypes of RUNX3 deficiency include expansion of the LSK+ HSPC compartment, and increased HSPC mobilization in response to G-CSF.22 Thus, loss of RUNX3 contributes to age-associated HSPC phenotypes, although likely co- operates with other perturbations to generate bona fide myeloid-skewing.
Additional studies have implicated RUNX3 in non-lym- phoid hematopoietic development. In human CD34+ cell erythroid cultures, RUNX3 was predicted by Cytoscape MiMI analysis of gene expression profiles to be a “parent protein,” i.e. a central node, in an erythroid transcription factor network.39 In zebrafish, morpholino knockdown of runx3 during embryogenesis resulted in severe anemia.19 Our results define a novel role for RUNX3 in the erythro- poietic program, governing the expression of lineage-spe- cific transcription factors such as KLF1, GATA1, and GFI1B. Notably, Klf1 and Gata1 displayed downregulation in aged murine HSC. We further show that RUNX3 defi- ciency produces perturbations at multiple developmental stages including MEP and committed erythroid progeni- tors. The decreased proliferation seen in RUNX3-deficient progenitors may contribute to differentiation impairment. However, the retained capacity for myeloid differentiation
AB
C
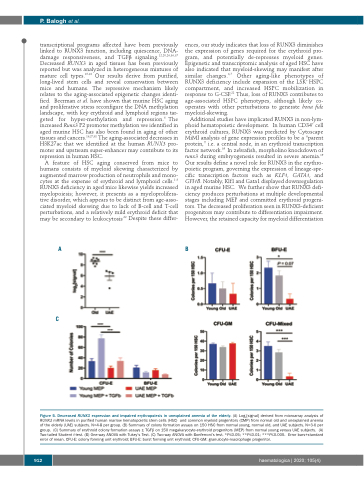
Figure 5. Decreased RUNX3 expression and impaired erythropoiesis in unexplained anemia of the elderly. (A) Log2[signal] derived from microarray analysis of RUNX3 mRNA levels in purified human marrow hematopoietic stem cells (HSC) and common myeloid progenitors (CMP) from normal old and unexplained anemia of the elderly (UAE) subjects. N=4-8 per group. (B) Summary of colony formation assays on 150 HSC from normal young, normal old, and UAE subjects. N=3-6 per group. (C) Summary of erythroid colony formation assays ± TGFβ on 150 megakaryocyte-erythroid progenitors (MEP) from normal young versus UAE subjects. (A) Two-tailed Student t-test. (B) One-way ANOVA with Tukey’s Test. (C) Two-way ANOVA with Bonferroni’s test. *P≤0.05; **P≤0.01; ***P≤0.005. Error bars+standard error of mean. CFU-E: colony forming unit erythroid; BFU-E: burst forming unit erythroid; CFU-GM: granulocyte-macrophage progenitor.
912
haematologica | 2020; 105(4)