Page 95 - 2020_02-Haematologica-web
P. 95
Irf2bp2b regulates zebrafish NMP cell fate choice
A
B
C D
E
F G
H
A’
B’
C’ D’
E’
F’ G’
H’
I
J
J’
K K’
L L’
N
O
M
P
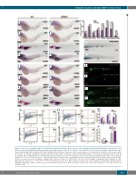
Figure 2. Deficiency of irf2bp2b leads to an expanded macrophage population at the expense of the neutrophil population during definitive myelopoiesis. (A-D’) Whole-mount in situ hybridization (WISH) analyses of neutrophil markers c/ebp1 (A, A’), mpx (B-C’), and lyz (D, D’) at 36 hours post-fertilization (hpf), 48 hpf, and 5 days post-fertilization (dpf) in wildtype (WT) and irf2bp2b-deficient embryos. Gray boxes and red arrows indicate the main position of positive cells for each marker. n/n, number of embryos showing representative phenotype/total number of embryos examined. (E-H’) WISH analyses of monocyte/macrophage markers csf1r (E, E’), mpeg1.1 (F, F’), and mfap4 (G-H’) at 48 hpf and 5 dpf. (I) Statistical results for A-H’. Error bars represent the mean ± standard deviation (SD) of at least 15-30 embryos. ***P<0.001 (Student t test). (J, J’) Sudan black-positive cells were reduced in irf2bp2b-deficient embryos at 3 dpf. (K, K’) Green fluorescence protein (GFP)- positive cells were decreased in irf2bp2b-/-//Tg(mpx:eGFP) embryos at 5 dpf. (L, L’) GFP-positive cells increased in irf2bp2b-/-//Tg(mpeg1.1:eGFP) embryos at 5 dpf. (M) Statistical results for J-L’. ***P<0.001; ****P<0.0001 (Student t test). (N, O) FACS analysis of eGFP-positive cells in wildtype and irf2bp2b-/-//Tg(mpx:eGFP) or irf2bp2b-/-//Tg(mpeg1.1:eGFP) embryos at 2 dpf. (P) Statistical results for N, O. Error bars represent the mean ± SD of three replicates. **P<0.01; ***P<0.001 (Student t test).
haematologica | 2020; 105(2)
329