Page 31 - Haematologica May 2022
P. 31
lncRNA variants in CN-AML
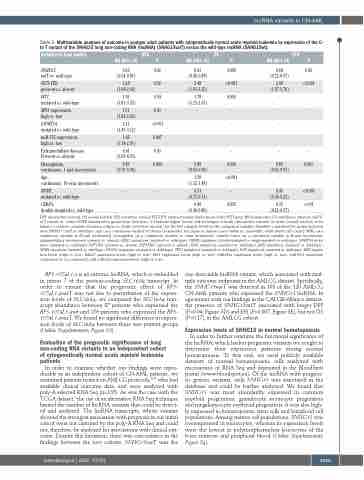
Table 2. Multivariable analyses of outcome in younger adult patients with cytogenetically normal acute myeloid leukemia by expression of the C- to-T variant of the SNHG15 long non-coding RNA (lncRNA) (SNHG15varT) versus the wild-type lncRNA (SNHG15wt).
Variables in final models
DFS OS EFS HR(95%CI) P HR(95%CI) P HR(95%CI) P
SNHG15, 0.63 varT vs. wild-type (0.34-0.81)
FLT3-ITD, 1.68 present vs. absent (1.08-2.60)
WT1, 1.91 mutated vs. wild-type (1.01-3.55)
MN1expression, 1.51 high vs. low (1.04-2.20)
0.02 0.63 0.008 0.68 0.02 (0.45-0.89) (0.22-0.57)
0.02 2.48 <0.001 2.08 (1.83-3.35) (1.57-2.76)
<0.001
0.04 1.78 0.004 - - (1.21-2.63)
0.03 - - - -
DNMT3A, 2.11 <0.001 mutated vs. wild-type (1.43-3.12)
miR-155 expression, 1.85 0.007 high vs. low (1.18-2.91)
Extramedullary disease, 0.61 0.03 Present vs. absent (0.39-0.95)
Hemoglobin, 0.87 0.004 continuous, 1-unit increments (0.79-0.96)
Age, -- continuous, 10-year increments
NPM1, - - mutated vs. wild-type
CEBPA, - - double-mutated vs. wild-type
-- - -
-- - -
-- - -
0.89 0.004 0.89 0.001 (0.83-0.96) (0.83-0.95)
1.29 <0.001 - - (1.12-1.48)
0.51 - 0.45 (0.37-0.71) (0.34-0.62)
0.49 0.005 0.35 (0.30-0.80) (0.22-0.57)
<0.001 <0.01
DFS: disease-free survival; OS: overall survival; EFS: event-free survival; FLT3-ITD: internal tandem duplications of the FLT3 gene; HR: hazard ratio; CI: confidence intervals; varT: C- to-T variant; vs.: versus. NOTE: Hazard ratios greater than (less than) 1.0 indicate higher (lower) risk for relapse or death (disease-free survival) or death (overall survival) or for failure to achieve complete remission, relapse or death (event-free survival) for the first category listed for the categorical variables.Variables considered for model inclusion were SNHG15 (varT vs. wild-type), age (as a continuous variable, in 10-year increments), sex (male vs. female), race (white vs. non-white), white blood cell count [(WBC) as a continuous variable, in 50-unit increments], hemoglobin (as a continuous variable, in 1-unit increments), platelet count (as a continuous variable, in 50-unit increments), extramedullary involvement (present vs. absent),ASXL1 mutations (mutated vs. wild-type),CEBPA mutations (double-mutated vs. single-mutated or wild-type),DNMT3A muta- tions (mutated vs. wild-type), FLT3-ITD (present vs. absent), FLT3-TKD (present vs. absent), IDH1 mutations (mutated vs. wild-type), IDH2 mutations (mutated vs. wild-type), NPM1 mutations (mutated vs. wild-type),RUNX1 mutations (mutated vs. wild-type),TET2 mutations (mutated vs. wild-type),WT1 mutations (mutated vs. wild-type),ERG expres- sion levels (high vs. low), BAALC expression levels (high vs. low), MN1 expression levels (high vs. low), miR-181a expression levels (high vs. low), miR-3151 expression (expressed vs. not expressed), and miR-155 expression levels (high vs. low).
RP5-1074L1.4 is an intronic lncRNA, which is embedded in intron 7 of the protein-coding SLC16A4 transcript. In order to ensure that the prognostic effect of RP5- 1074L1.4varT was not due to perturbation of the expres- sion levels of SLC16A4, we compared the SLC16A4 tran- script abundance between 87 patients who expressed the RP5-1074L1.4wt and 156 patients who expressed the RP5- 1074L1.4varT. We found no significant difference in expres- sion levels of SLC16A4 between these two patient groups (Online Supplementary Figure S3).
Evaluation of the prognostic significance of long non-coding RNA variants in an independent cohort of cytogenetically normal acute myeloid leukemia patients
In order to examine whether our findings were repro- ducible in an independent cohort of CN-AML patients, we examined patients treated on AMLCG protocols,38,39 who had available clinical outcome data, and were analyzed with poly-A selected RNA Seq (n=135). As was the case with the TCGA dataset,8 the use of an alternative RNA Seq technique limited the number of lncRNA variants that could be detect- ed and analyzed. The lncRNA transcripts, whose variants showed the strongest association with prognosis in our initial cohort were not captured by the poly-A RNA Seq and could not, therefore, be analyzed for associations with clinical out- come. Despite this limitation, there was concordance in the findings between the two cohorts. SNHG15varT was the
one detectable lncRNA variant, which associated with mul- tiple outcome endpoints in the AMLCG dataset. Specifically, the SNHG15varT was detected in 103 of the 120 AMLCG CN-AML patients who expressed the SNHG15 lncRNA. In agreement with our findings in the CALGB/Alliance dataset, the presence of SNHG15varT associated with longer DFS (P=0.04; Figure 3D) and EFS (P=0.007, Figure 3E), but not OS (P=0.17), in the AMLCG cohort.
Expression levels of SNHG15 in normal hematopoiesis In order to further examine the functional significance of the lncRNA, which harbor prognostic variants we sought to determine their expression patterns during normal hematopoiesis. To this end, we used publicly available datasets of normal hematopoietic cells analyzed with microarrays or RNA Seq and deposited in the BloodSpot portal (www.bloodspot.eu). Of the lncRNA with prognos- tic genetic variants, only SNHG15 was annotated in the database and could be further analyzed. We found that SNHG15 was most abundantly expressed in common myeloid progenitors, granulocyte monocyte progenitors and megakaryocyte-erythroid progenitors. It was also high- ly expressed in hematopoietic stem cells and lymphoid cell populations. Among mature cell populations, SNHG15 was overexpressed in monocytes, whereas its expression levels were the lowest in polymorphonuclear leucocytes of the bone marrow and peripheral blood (Online Supplementary
Figure S4).
haematologica | 2022; 107(5)
1039