Page 28 - Haematologica May 2022
P. 28
D. Papaioannou et al.
Multivariable proportional hazards models were constructed using a backward selection procedure.45 All statistical analyses using CALGB/Alliance data were performed by the Alliance Statistics and Data Center.
In vitro experiments
LncRNA wild-type and variant transcripts were amplified with
Phusion high fidelity polymerase by polymerase chain reaction (PCR). Amplicons were cloned into pcDNA using the Gibson technique, according to standard protocols. For primer sequences and further experimental details please see the Online Supplementary Appendix. K-562 and THP-1 cells were transfected with vectors containing either a cytosine (C)-to-thymidine (T) variant in the lncRNA SNHG15 (SNHG15varT) or wild-type lncRNA SNHG15 (SNHG15wt); cells were also transfected with empty pcDNA3.1 and were used as controls. Cell viability and apoptosis were assessed with annexin V staining. The colorimetric MTT assay was used to assess the proliferative capacity of the transfected blasts.
Results
Detection of genetic variants in the non-coding transcriptome of younger adults with cytogenetically normal acute myeloid leukemia
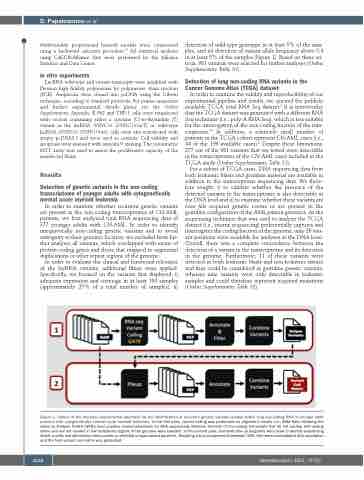
In order to examine whether recurrent genetic variants are present in the non-coding transcriptomes of CN-AML patients, we first analyzed total RNA sequencing data of 377 younger adults with CN-AML. In order to identify unequivocally non-coding genetic variants and to avoid ambiguity in their genomic location, we excluded from fur- ther analyses all variants, which overlapped with exons of protein-coding genes and those that mapped to segmental duplications or other repeat regions of the genome.
In order to evaluate the clinical and functional relevance of the lncRNA variants, additional filters were applied. Specifically, we focused on the variants that displayed: i) adequate expression and coverage in at least 100 samples (approximately 25% of a total number of samples), ii)
detection of wild-type genotype in at least 5% of the sam- ples, and iii) detection of variant allele frequency above 0.4 in at least 5% of the samples (Figure 1). Based on these cri- teria, 981 variants were selected for further analyses (Online Supplementary Table S1).
Detection of long non-coding RNA variants in the Cancer Genome Atlas (TCGA) dataset
In order to examine the validity and reproducibility of our experimental pipeline and results, we queried the publicly available TCGA total RNA Seq dataset.8 It is noteworthy that the TCGA dataset was generated with a different RNA Seq technique (i.e., poly-A RNA Seq), which is less suitable for the interrogation of the non-coding fraction of the tran- scriptome.46 In addition, a relatively small number of patients in the TCGA cohort represent CN-AML cases (i.e., 44 of the 196 available cases).8 Despite these limitations, 277 out of the 981 variants that we tested were detectable in the transcriptomes of the CN-AML cases included in the TCGA study (Online Supplementary Table S2).
For a subset of TCGA cases, DNA sequencing data from both leukemic blasts and germline material are available in addition to the transcriptome sequencing data. We there- fore sought: i) to validate whether the presence of the detected variants in the transcriptome is also detectable at the DNA level and ii) to examine whether these variants are bona fide acquired genetic events or are present in the germline configuration of the AML patient genomes. As the sequencing technique that was used to analyze the TCGA dataset (i.e., exome sequencing) preferentially captures and interrogates the coding fraction of the genome, only 20 vari- ant positions were available for analyses at the DNA level. Overall, there was a complete concordance between the detection of a variant in the transcriptome and its detection in the genome. Furthermore, 11 of these variants were detected in both leukemic blasts and non-leukemic tissues and thus could be considered as germline genetic variants, whereas nine variants were only detectable in leukemic samples and could therefore represent acquired mutations (Online Supplementary Table S2).
Figure 1. Outline of the two-pass experimental approach for the identification of recurrent genetic variants located within long non-coding RNA in younger adult patients with cytogenetically normal acute myeloid leukemia. In the first pass, variant calling was performed on alignment results (i.e., BAM files) following the Genome Analysis Toolkit (GATK) best practice recommendations for RNA sequencing datasets. Variants of non-coding transcripts that do not overlap with coding exons and are not located in low-complexity regions of the genome were selected. In the second pass, Samtools pile-up programs were used to identify sequencing depth, quality and alternative allele counts on selected unique variant positions. Resulting visual component framework (VCF) files were consolidated with annotation and the final variant call matrix was generated.
1036
haematologica | 2022; 107(5)