Page 210 - 2022_03-Haematologica-web
P. 210
Letters to the Editor
tologic lineages, including lymphocyte subsets, were within their normal ranges. These four patients had high levels of circulating IgG and/or IgA at diagnosis which persisted throughout follow-up.
We investigated a possible genetic etiology using target- ed sequencing of genes known to be involved in inherited neutropenia and exome-sequencing. We excluded CXCR4
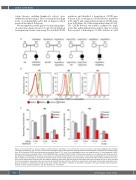
mutations and identified a homozygous CXCR2-gene deletion in P1, homozygous CXCR2 missense mutations in P2 and P3, and compound heterozygous CXCR2 muta- tions in P4 (Figure 1A, Online Supplementary Figure S1A, B). The CXCR2 deletion was further confirmed by single nucleotide polymorphism-array analysis (data not shown) that revealed a homozygous 13.4-kb deletion in 2q35
A
B
C
Figure 1. Characterization of germline biallelic CXCR2 mutations identified in four patients with chronic neutropenia. (A) Family pedigrees with identified homozygous (patients P1, P2, and P3) or compound heterozygous (P4) CXCR2 mutations. Healthy parents were heterozygous carriers for the identified muta- tions. (B) Cell-surface CXCR2 immunostaining on neutrophils from P1, P2, and P3, one heterozygous carrier, and healthy donors. (C) Dose-dependent CXCL8- induced chemotaxis of neutrophils without or with SB265610 (SB), its specific CXCR2 inhibitor. Chemotaxis assays were run in duplicate, with whole blood sam- ples (diluted 1:4 in RPMI with 1% human serum) using 12 mm diameter transwell devises with 5 mm pores. For each assay including patient, parent and control, blood samples were collected concomitantly and treated equally. Samples were added in the upper chamber, CXCL8 in the lower chamber and SB in both cham- bers. Control wells without chambers were also added to determine the number and phenotype of total seeded cells. After incubation for 1 hour, cells recovered in the lower chambers (responding cells) were counted and identified by flow cytometry. Results are expressed as percentage of responding neutrophils, calcu- lated as [(Number of neutrophils recovered in the lower chamber with CXCL8) - (Number of neutrophils recovered in the lower chamber without CXCL8)] / (Number of total seeded neutrophils) x 100. WT: wild-type; na: not available.
766
haematologica | 2022; 107(3)