Page 238 - 2021_03-Haematologica-web
P. 238
Letters to the Editor
A
BD
C
F
E
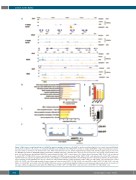
Figure 3. NA9 induces a rapid modification of H3K27ac marks in multiple enhancers. (A) H3K27ac peaks on indicated hg19 loci in control- (top) and NA9 (mid- dle)-transduced CD34+ cells, in blue, as compared with NA9 peaks identified in the HEK293FT cell line, in orange (GSE62587).3 A green bar indicates an 11.7 kb super enhancer found 10.7 kb downstream of the PBX3 transcriptional start site (Online Supplementary Dataset S9). (B) Most significant gene sets from the MySigDB perturbation, MySigDB pathway and Panther pathway ranked by a binomial false discovery rate q-value (blue). Percentage overlap between the gene sets and the genes with proximal H3K27ac marks identified in NA9+ (GFP+) chronic phase (CP) chronic myeloid leukemia (CML) CD34+ cells and absent in control (YFP+) CP CML CD34+ cells are also indicated (orange). (C) H3K27ac peaks present in NA9+ (GFP+) CD34+ cells and absent in control (YFP+) cells over- lap with enhancers of genes upregulated in monocytes, macrophages, neutrophils and dendritic cells following exposure to lipopolysaccharides. (D) Transcription factors showing enriched binding sites in gene enhancers with proximal H3K27ac marks identified in NA9+ (GFP+) CP CML CD34+ cells and absent in control (YFP+) CP CD34+ cells. (E) Average number of super enhancers found in control (YFP+) CP CML CD34+ cells only (CTR), or NA9+ (GFP+) CP CML CD34+ cells only (NA9), or in both NA9+ and control cells (CTR & NA9). Differences were not statistically significant (Wilcoxon matched-pairs signed ranked test). (F) Location of two super enhnacers (gray bars) found within the ANGPT1 gene in NA9+ (GFP+) CP CML CD34+ cells and not in control (YFP+) CP CML CD34+ cells.
884
haematologica | 2021; 106(3)