Page 25 - Haematologica-April 2018
P. 25
IKZF1 in leukemia and therapy response
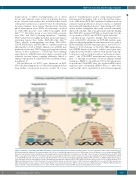
translocations or CRLF2 rearrangements, act as driver lesions and represent early events in leukemia develop- ment. Genome-wide analysis has established that several other genetic alterations cooperate before B-cell leukemia becomes manifest. Gene lesions that inactivate the lym- phoid transcription factor IKZF1 are frequently observed in BCR-ABL1-positive and CRLF2-rearranged BCP- ALL.65,69,91,92 The latter group is associated with concomi- tant JAK1 and JAK2 activating mutations.91 Similarly, IKZF1 alterations are highly prevalent in tyrosine kinase- activating lesions that define BCR-ABL1-like ALL.67,68 These include rearrangements involving ABL1/ABL2, CSF1R, EPOR, JAK2 and PDGFRB, or sequence mutations affecting FLT3, IL7R or SH2B3. Indeed, loss of IKZF1 may permit more effective STAT5 target gene regulation down- stream of these pathways.93 Collectively, these findings argue that loss of IKZF1 function strongly cooperates with activated tyrosine kinase signaling pathways linked to enhanced progenitor B-cell proliferation and immortaliza- tion (Figure 3).
The predilection for IKZF1 gene alterations in BCR- ABL1-mediated lymphoid versus myeloid malignancies has been further corroborated in mouse studies. In a bone
marrow transplantation model using lineage-negative hematopoietic progenitor cells, it was shown that expres- sion of IK6 skews BCR-ABL1-mediated leukemia from an exclusive myeloproliferative disease towards a combined myeloid and B-lymphoid disease.63 Introducing p19Arf-defi- ciency further strengthens this trend towards uniformly induced B-cell ALL. This is in agreement with the finding that BCR-ABL1-positive BCP-ALL is characterized by the co-occurrence of IKZF1 and CDKN2A gene deletions.65
Another group of genetic changes that frequently co- occur with IKZF1 alterations in BCP-ALL include gene deletions affecting lymphoid transcription factors, such as EBF1 and PAX5, and the transcriptional co-factor BTG148,58 (Figure 3). BTG1 belongs to the BTG/TOB antiprolifera- tive (APRO) family of proteins,94 which control gene tran- scription by their ability to interact with specific transcrip- tion factors, such as nuclear receptors and homeobox pro- teins.95,96 the CCR4-NOT transcriptional regulatory com- plex,97 or through recruitment of protein arginine methyl transferase PRMT1.98 In addition, BTG1 through interac- tion with the CCR4-NOT, may also regulate mRNA dead- enylation and consequently mRNA decay.99,100 Mice defi- cient for Btg1 show a partial block in B-cell development,
Figure 3. Pathways cooperating with IKZF1 alterations in leukemia pathogenesis. Pathways involving cytokine receptor signaling and B-cell differentiation by lym- phoid transcriptional regulators in normal progenitor B cells are schematically indicated on the left. Alterations of these pathways co-occur frequently with IKZF1 deletions and mutations in B-cell progenitor acute lymphoblastic leukemia (BCP-ALL) as indicated on the right. These include, activating mutations in FLT3, IL7R, JAK2 (*), upregulation of CRLF2 (+), C-terminal truncations or upregulation of EPOR (^), chromosomal translocations generating fusion proteins with PDGFR or CSF1R (-), and BCR-ABL1, which collectively results in activated cytokine receptor and tyrosine kinase signaling leading to STAT activation. In addition, IKZF1 alterations co- occur with gene deletions affecting the activity of B-lymphoid transcriptional regulators EBF1, PAX5 and BTG1, which results in a block of B-cell differentiation. FLT3: FMS related tyrosine kinase 3; IL7R: interleukin 7 receptor; CRLF2: cytokine receptor like factor 2; C-KIT: mast/stem cell growth factor receptor Kit; JAK, Janus kinase; STAT: signal transducer and activator of transcription; BTG1: B-cell translocation gene 1; EBF1: early B-cell factor 1; PAX5: paired box 5; IKZF1: IKAROS family zinc finger 1; CSF1R: colony-stimulating factor 1 receptor; EPOR: erythropoietin receptor; PDGFR: platelet-derived growth factor receptor.
haematologica | 2018; 103(4)
569