Page 21 - Haematologica May 2020
P. 21
Editorials
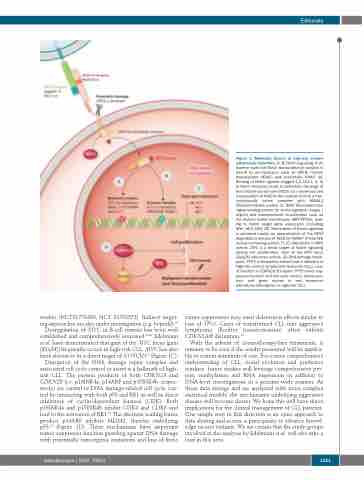
studies (NCT01778439, NCT 01703572). Indirect target- ing approaches are also under investigation (e.g. bepridil).38 Dysregulation of MYC in B-cell tumors has been well established and comprehensively reviewed.39,40 Edelmann et al. have demonstrated that gain of the MYC locus [gain (8)(q24)] frequently occurs in high-risk CLL. MYC has also been shown to be a direct target of NOTCH141 (Figure 1C). Disruption of the DNA damage repair complex and associated cell cycle control or arrest is a hallmark of high- risk CLL. The protein products of both CDKN2A and CDKN2B (i.e. p16INK4a, p14ARF and p15INK4b, respec- tively) are central to DNA damage-related cell cycle con- trol by interacting with both p53 and RB1 as well as direct inhibitors of cyclin-dependent kinases (CDK). Both p16INK4a and p15INK4b inhibit CDK4 and CDK6 and lead to the activation of RB1.42 The alternate reading frame product p14ARF inhibits MDM2, thereby stabilizing p53.43 (Figure 1D). These mechanisms have important tumor suppressor function guarding against DNA damage with potentially tumorigenic mutations and loss of these
Figure 1. Molecular drivers of high-risk chronic lymphocytic leukemia. (A, B) Notch signaling. In its inactive state the Notch transcriptional complex is bound by co-repressors such as SPEN, histone deacetylases (HDAC) and, potentially, SNW1 (A). Binding of Notch ligands (Jagged-1,2, DLL1, 3, 4) to Notch receptors leads to proteolytic cleavage of the intracellular domain (NICD) via γ-secretase and translocation of NICD to the nucleus to form a tran- scriptionally active complex with MAML1 (Mastermind-like protein 1), RBPJ (Recombination signal binding protein for immunoglobulin kappa J region) and transcriptional co-activators such as the histone acetyl transferases CBP/EP300, lead- ing to Notch target gene expression (including MYC, HES, HEY) (B). Termination of Notch signaling is achieved mainly via ubiquitination of the PEST degradation domain of NICD by FBXW7 (F-box/WD repeat-containing protein 7). (C) Alterations in MYC activity. MYC is a direct target of Notch signaling driving cell proliferation. Gain of the MYC locus (8)(q24) enhances activity. (D) DNA damage check- point. TP53 is frequently altered and a hallmark of high-risk chronic lymphocytic leukemia (CLL). Loss of function in CDKN2A/B impairs TP53 tumor sup- pressor function and cell cycle control. (Gene sym- bols and gene names in red represent altered/mutated genes in high-risk CLL).
tumor suppressors may exert deleterious effects similar to loss of TP53. Cases of transformed CLL into aggressive lymphoma (Richter transformation) often exhibit CDKN2A/B disruption.44
With the advent of chemotherapy-free treatments, it remains to be seen if the results presented will be applica- ble to current standards of care. For a more comprehensive understanding of CLL, clonal evolution and predictive markers, future studies will leverage comprehensive pro- tein, methylation and RNA expression in addition to DNA-level investigations in a genome-wide manner. As these data emerge and are analyzed with more complex statistical models7 the mechanisms underlying aggressive disease will become clearer. We hope this will have direct implications for the clinical management of CLL patients. One simple step in this direction is an open approach to data sharing and access, a prerequisite to advance knowl- edge on rare variants. We are certain that the study groups involved in the analysis by Edelmann et al. will also take a lead in this area.
haematologica | 2020; 105(5)
1181