Page 276 - 2020_02-Haematologica-web
P. 276
L. Martorell et al.
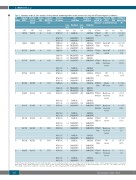
Table 2. Summary of the in silico analysis of the predicted readthrough effect at the protein level using the SIFT and Polyphen-2 softwares.
F8 Mutation Mutation Affected Predicted Predicted
SIFT prediction
Polyphen-2 Sequence Expected FVIII:C FVIII:Ag(% prediction context/RT effect (fold before/after
variant HGVS* Legacy† FVIII MW (kDa)/ domain (%)‡
0 WT WT N.A 166.19
AA change
efficiency# on protein increase) RT
1 W274X
2 Q462X
3 Q1705X
4 R1715X
5 W1726X
6 Q1764X
7 R1822X
8 R1960X
9 W2015X
10 R2071X
11 W2131X
12 R2228X
W255X A1
Q443X A2
Q1686X A3
R1696X A3
W1707X A3
Q1745X A3
R1803X A3
R1941X A3
W1996X A3
R2052X C1
W2112X C1
R2209X C2
30.92
52.8
91.66
92.85
94.21
98.51
105.24
121.5
127.9
134.11
140.99
151.85
W/W/(98)
W/R/(1.2)
W/C/(0.4) Q/Q/(64) Q/K/(34) Q/L/(0.5) Q/Q/(64)
Q/K/(34) Q/L/(0.5) R/W/(98)
R/R/(1.2)
R/C/(0.4) W/W/(98)
W/R/(1.2) W/C/(0.4) Q/Q/(64)
Q/K/(34)
Q/L/(0.5) R/W/(98)
R/R/(1.2) R/C/(0.4) R/W/(98)
R/R/(1.2)
R/C/(0.4) W/W/(98)
W/R/(1.2) W/C/(0.4) R/W/(98)
R/R/(1.2)
R/C/(0.4) W/W/(98)
W/R/(1.2) W/C/(0.4) R/W/(98)
R/R/(1.2)
R/C/(0.4)
1 SAME AA 0
0 DAMAGING 1
0 DAMAGING 1 1 SAMEAA 0
SAME AA
DAMAGING
DAMAGING SAME AA DAMAGING DAMAGING SAME AA
DAMAGING BENIGN DAMAGING
SAME AA
DAMAGING SAME AA
DAMAGING DAMAGING SAME AA
BENIGN
BENIGN DAMAGING
SAME AA DAMAGING DAMAGING
SAME AA
DAMAGING SAME AA
DAMAGING DAMAGING DAMAGING
SAME AA
DAMAGING SAME AA
DAMAGING DAMAGING DAMAGING
SAME AA
DAMAGING
T TGA C/
Highest
T TAG C/ High
T TAG A/
Low
ATGAC/
High
CTGAG/ Low
TTAGC/
High
WT 3.7
variant
Mainly WT 2.6 variant
Mainly WT 1
variant
Mainly non 1.2
functional variant
WT 1 variant
Mainly 3.3
WT variant
1.55 / 24.6
(15.87)
3.65 / 18.7 (5.12)
1.25 / 4.5
(3.6)
0.6 / 6.7
(11.17)
0.75 / 5.0 (6.67)
1.6 / 13.4
(8.37)
0.4 / 5.3 (13.25)
1.1 / 9.55
(8.68)
0.5 / 15.65 (31.3)
0.35/7
(20)
2.85 / 13 (4.56)
0.6/3
(5)
Score Predicted Score Predicted [0-1] [1-0]
N.A N.A N.A N.A N.A ATGAC N.A N.A N.A
0.26 0.02 1
0.37 0.37 0.05
1
0 1
0 0 1
0.67
0.13 0.04
1 0.01 0
1
0 1
0.01 0 0
1
0 1
0 0 0
1
0
TOLERATED 0.99 DAMAGING 0.99 SAME AA 0
TOLERATED 0.99 TOLERATED 0.397
DAMAGING
SAME AA
DAMAGING SAME AA
DAMAGING DAMAGING SAME AA
1
0
1 0
1 1 0
TOLERATED 0.062
TOLERATED 0.012 DAMAGING 0.887
SAME AA 0 DAMAGING 0.84
Mainly non 1.3 variant
TTGAT/ Mainly non 1
Medium functional variant
DAMAGING
SAME AA
DAMAGING SAME AA
DAMAGING DAMAGING DAMAGING
SAME AA
DAMAGING SAME AA
DAMAGING DAMAGING DAMAGING
SAME AA
DAMAGING
1
0
1 0
1 1 1
0
1 0
1 1 1
0
1
TTGAC/ Highest
CTGAC/
High
GTGAC/ High
TTGAC/
WT 2.9 variant
Mainly non 1.2
functional variant
WT 2.7 variant
Mainly non 1.1
TTGAA/
Medium functional
Highest functional variant
*HGVS:Human GenomeVariation Society numbering,where codons are numbered with codon +1 coding for the first residue (Met) of the 19-residue signal peptide (this is -19 in Legacy numbering). †Legacy numbering, where codon +1 refers to that encoding the first AA of the mature FVIII protein (in HGVS numbering, it is codon +20). ‡According to Roy et al.15. #According to nucleotide context rule (-1-Stop-+4).AA:amino acid.N.A:not aplicable.RT:readthrough.WT:wild type.
510
haematologica | 2020; 105(2)