Page 114 - 2019_06-Haematologica-web
P. 114
C. Bueno et al.
tions affected by genes differentially expressed in MA4-, A4M- and double fusion-expressing HEP relative to EV were classified by Ingenuity Pathway Analysis soft- ware;47,52 the top significantly enriched functional cate- gories included “hematological system development and function”, “cancer” and “hematological disease” (Figure 6B). Statistical (-logp-value) power shows distant effects of MA4 and A4M; however, co-expression of both fusions seems to establish a molecular balance/developmental cooperation in promoting blood-endothelial specification from hPSC. Strikingly, biofunctions specifically associat- ed with hematologic lymphoid malignancies (not with non-hematologic cancer) were predicted to be activated (positive z-score) exclusively in double fusion-expressing HEP, further suggesting a molecular cooperation between MA4 and A4M in development and infant leukemia. (Figure 6C).
The C-terminal partners of MLL fusions normally inter- act with the histone methyltransferase DOT1L, which is the sole histone methyltransferase catalyzing histone 3 lysine 79 (H3K79) methylation, a chromatin modification widely associated with the dysregulated expression of HOX-A cluster genes in MLL leukemias.13,53 We thus per- formed genome-wide chromatin immunoprecipitation-
A
BD
sequencing analysis of the H3K79 trimethylation (H3K79me3) profiles in control, MA4-, A4M- and double fusion-expressing hESC-derived blood derivatives (Figure 7, Online Supplementary Figure S3A, Online Supplementary Table S3). In line with the RNA-sequencing data, function- al analysis of the differential H3K79me3 peaks specific for double fusion-expressing cells revealed significant gene ontology functional categories associated with ”definitive hematopoiesis”, “myeloid and erythroid differentiation/homeostasis” and “endothelial cell devel- opment” (Figure 7A, Online Supplementary Figure S3B). This further supports the developmental co-operation between A4M and MA4 in promoting hemato-endothelial specification.
Finally, we analyzed the H3K79me3 profiles at genomic loci of the classical MLL target genes reported by Guenther et al.54 Non-HOX-A classical MLL targets such as RUNX1, LMO2, ADMA10, and KDM6A showed a slight although non-significant, enrichment of H3K79me3 in MA4-expressing hESC, validating our chromatin immunoprecipitation-sequencing approach (Figure 7B). However, enrichment of H3K79me3 in HOX-A cluster genes was observed exclusively in A4M-expressing cells although it was statistically significant only in double
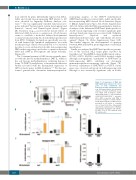
Figure 5. Co-expression of MA4 and A4M significantly enhances the emer- gence of both endothelial and hemogenic hemato-endothelial precur- sors. (A) Representative flow cytometry analysis of hemato-endothelial precur- sors (HEP) with hemogenic (CD45- CD31+CD43+CD34dim/+) and endothelial (CD45-CD31+CD43-CD34++) potential. (B,C) A4M co-operates with MA4 to boost the emergence of both endothe- lial (B) and hemogenic (C) HEP. Data are presented as the mean ± standard error of mean from three independent experiments. (D) Expression of RUNX1c and Ve-Cad in hemogenic and endothe- lial HEP. *P<0.05. EB: erythroid body; EV: empty vector.
C
1196
haematologica | 2019; 104(6)